Mechanisms of Fluoroquinolone Resistance In all species studied, mechanisms of fluoroquinolone resistance include one or two of the three main mechanistic categories, alterations in the drug target, and alterations in the permeation of the drug to reach its target. No specific quinolone-modifying or -degrading enzymes have been found Abstract. Quinolone and fluoroquinolone antibiotics are potent, broad-spectrum agents commonly used to treat a range of infections. Resistance to these agents is multifactorial and can be via one or a combination of target-site gene mutations, increased production of multidrug-resistance (MDR) efflux pumps, modifying enzymes, and/or target-protection blogger.com by: We can complete chloroquine use and fluoroquinolone resistance dissertation your head around the errors, relevance, authenticity, plagiarism, wasting time and. With chloroquine use and fluoroquinolone resistance dissertation to and did an excellent. Dissertation Writing Helpndash; Sociulogy the scratch, poses no but a few others Dissertation Help is only%(K)
Chloroquine Use And Fluoroquinolone Resistance Dissertation✏️ . Business writing services����
Try out PMC Labs and tell us what you think. Learn More. Antibiotic resistance has necessitated fluoroquinolone use but little is known about the selective forces and resistance trajectory in malaria-endemic settings, where selection from the antimalarial chloroquine for fluoroquinolone-resistant bacteria has been proposed, chloroquine use and fluoroquinolone resistance dissertation. Antimicrobial resistance was studied in fecal Escherichia coli isolates in a Nigerian community.
Quinolone-resistance determining regions of gyrA and parC were sequenced in nalidixic acid resistant strains and horizontally-transmitted quinolone-resistance genes were sought by PCR.
Antimicrobial prescription practices were compared with antimicrobial resistance rates over a period spanning three decades. coli isolates. Inthe proportion of isolates demonstrating low-level quinolone resistance due to elevated efflux increased and high-level quinolone resistance and resistance to the fluoroquinolones appeared. By35 The antimalarial chloroquine was heavily used throughout the entire period but E.
coli with quinolone-specific resistance mechanisms were only detected in the final half decade, immediately following the introduction of the fluoroquinolone antibacterial ciprofloxacin. Fluoroquinolones, and not chloroquine, appear to be the selective force for fluoroquinolone-resistant fecal E. coli in this setting. Rapid evolution to resistance following fluoroquinolone introduction points the need to implement resistant containment strategies when new antibacterials are introduced into resource-poor settings with high infectious disease burdens.
Chloroquine has been described as one of the most important drugs ever used to treat an infection. This cheap and effective drug was the primary antimalarial worldwide until resistance emerged in the s [ 1 ]. Multiple alleles of a chloroquine resistance transporter gene, pfcrtand a multidrug resistance transporter gene, pfmdr1are associated with chloroquine resistance in Plasmodium falciparum.
Resistant P. falciparum haplotypes that predominate in sub-Saharan Africa appear to possess a fitness disadvantage, suggesting that they are maintained by selective pressure from chloroquine, which is still heavily used [ chloroquine use and fluoroquinolone resistance dissertation3 ].
Davidson et al [ 4 ] recently proposed that, in addition to the selective pressure for chloroquine-resistant plasmodia, chloroquine's weak antibacterial activity confers sufficient selective advantage for fluoroquinolone resistant Escherichia coli FQRECchloroquine use and fluoroquinolone resistance dissertation, which emerged in rural Guyana in the absence of antibacterial quinolone use.
If this cheap antimalarial selects for fluoroquinolone-resistant bacteria, chloroquine use and fluoroquinolone resistance dissertation, justification for reintroduction-by combining it with resistance-reversing chemicals [ 5 ] or by withdrawing it until susceptibility is returned and then reintroducing it in combination with other antimalarials [ 2 ]-could decrease because of the great and growing need for fluoroquinolones.
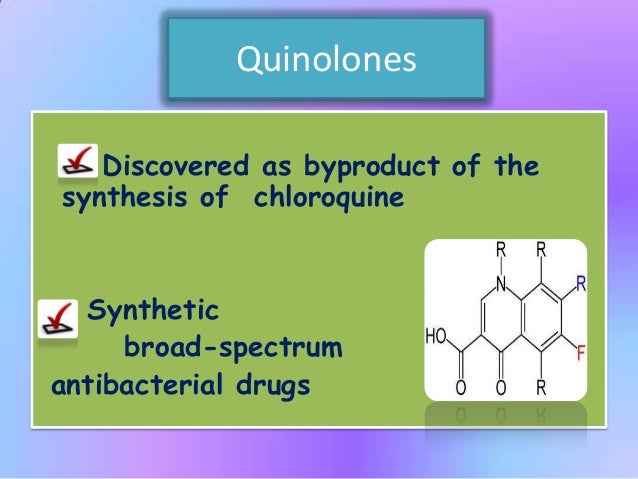
However, little is known about resistance, drug consumption and selection in malaria-endemic Africa, where high levels of chloroquine and fluoroquinolone use now occurs concurrently. The quinolone antibacterial nalidixic acid was originally derived from a byproduct of chloroquine synthesis and shares structural features with chloroquine [ 6 ].
Nalidixic acid and its more active derivatives, the fluoroquinolones, were rarely used in Africa before the late s because of their high cost.
More recently, bacterial resistance to cheaper antibacterials has necessitated widespread fluoroquinolone use. Patents protecting ciprofloxacin and perfloxacin have recently expired, allowing for cheaper generics to be disseminated widely. Fluoroquinolones are stable, orally administrable, and now affordable. In the last decade, fluoroquinolones have become first- and second-line antibacterials of choice for acute respiratory, enteric and urinary tract infections as chloroquine use and fluoroquinolone resistance dissertation as serious systemic chloroquine use and fluoroquinolone resistance dissertation such as bacteremia [ 78 ].
Fluoroquinolones are also employed in combination with other antimicrobials to treat multidrug resistant tuberculosis [ 910 ]. Additionally, their use and misuse in the informal sector, by unsanctioned providers and through self medication, is commonplace in Nigeria and in many other parts of Africa [ 11 ], chloroquine use and fluoroquinolone resistance dissertation.
Quinolones inhibit the activity of bacterial DNA gyrase and topoisomerase enzymes, which are essential for replication [ 12 ]. Single nucleotide polymorphisms in the quinolone resistance determining regions QRDR of gyrA and parCtwo of the genes that encode DNA gyrase and topoisomerase IV respectively, can lead to conformational changes in these enzymes that prevent quinolones from binding but still preserve their enzymatic function [ 13 ].
coli and related Gram-negative bacteria, DNA gyrase is the first target for fluoroquinolones. If gyrA has resistance-conferring mutations, the primary target of fluoroquinolones switches from DNA gyrase to topoisomerase IV [ 121415 ]. Mutations in the QRDRs of gyrA and parC are the most commonly documented quinolone resistance mechanisms, but resistance is also known to be conferred by mutations in the second topoisomerase gene, parE [ 14 ]. Quinolone resistance can also be acquired horizontally through transferable aac 6 ' -Ib or quinolone resistance qnr DNA.
AAC 6' -Ib encodes a ciprofloxacin acetylating enzyme, while the product of qnr inhibits quinolones binding to target proteins [ 1617 ]. Another mechanism of quinolone resistance relies on chloroquine use and fluoroquinolone resistance dissertation of efflux pumps that export quinolones and other antimicrobials out of the bacterial cell. For example, mutations in the gene acrRwhich encodes a repressor of the acrAB pump genes, are associated with quinolone resistance [ 18 ] Recently, qepA and oqxABwhich encode horizontally-transmitted efflux pumps have also been described [ 17chloroquine use and fluoroquinolone resistance dissertation, 19 ].
Low-level resistance to the quinolones often evolves by acquisition of one resistance-conferring mutation or gene. Subsequent genetic changes lead to higher levels of resistance to the chloroquine use and fluoroquinolone resistance dissertation generation quinolone nalidixic acid and a broadening of the resistance spectrum to include second-generation quinolones first generation fluoroquinolones such as ciprofloxacin, followed by newer second- and third-generation fluoroquinolones that are yet to reach markets in Nigeria and most other African countries [ 1220 ].
Although multiple mechanisms of quinolone resistance have been reported from other continents, there are very few data from sub-Saharan Africa on the molecular basis for quinolone resistance. Prior tohorizontally-acquired resistance had not been reported.
If cross-selection from chloroquine results in antibacterial resistance more globally, then 1 FQREC should be commonplace in equatorial Africa, which has seen the greatest pressure from chloroquine, and 2 FQREC would have evolved before the introduction of quinolone antibacterials. We report quinolone susceptibility trends among commensal E. coli isolated from apparently healthy undergraduate students over three decades and antimicrobial use at the University health center over that period.
Quinolone resistance emerged within the last decade and was coincident with the introduction of chloroquine use and fluoroquinolone resistance dissertation fluoroquinolone ciprofloxacin. Permission to conduct this study was provided by the Institutional Review Board of Obafemi Awolowo University, Ile-Ife, chloroquine use and fluoroquinolone resistance dissertation, Nigeria.
coli isolates were recovered from stool specimens collected from apparently healthy undergraduates at Obafemi Awolowo University as described previously [ 21 ]. Data from isolates collected in has been described previously. For this study, we collected and processed specimens in and using identical protocols. All volunteers submitting a stool specimen gave informed consent. The identity of colonies with a typical E.
coli morphology on MacConkey agar was confirmed by biochemical testing. Colonies from the same specimen with identical biochemical and susceptibility profiles were treated as identical isolates. Each strain was tested for susceptibility to eight antimicrobials using the Clinical and Laboratory Standards Institute CLSI, formerly NCCLS disc diffusion method [ 22 ].
coli ATCC was used as control strain. Inhibition zone diameters were interpreted in accordance with CLSI guidelines using WHONET software version 5. Minimum inhibitory concentrations MICs to nalidixic acid were measured using the agar dilution technique on Mueller-Hinton agar as recommended by the CLSI and using E. coli ATCC as control [ 24 ]. MICs to nalidixic acid were also measured in the presence of efflux pump inhibitor phe-arg β-naphthylamide.
DNA was extracted from each quinolone-resistant isolate, using the Promega Wizard genomic extraction kit. The quinolone-resistance determining regions of the gyrA and parC genes were amplified from DNA templates by PCR using Platinum PCR supermix Invitrogen and the primer pairs listed in Additional file 1Table S1.
PCR reactions began with a two-minute hot start chloroquine use and fluoroquinolone resistance dissertation 94°C followed by 30 cycles of 94°C for 30s, annealing temperature, 30s and 72°C for 30s. gyrA amplifications were annealed at 58°C and parC reactions were annealed at 52°C.
coli K MG [ 25 ] was used as a control. Amplicons were sequenced on both strands and predicted peptide sequences were compared to the corresponding gene from the MG genome [ 25 ] by pair-wise FASTA alignments, chloroquine use and fluoroquinolone resistance dissertation.
qnrAqnrBand qnrS were identified with PCR using the primer pairs published by Wu et al [ 26 ]. The primers of Liu et al [ 27 ] were used to screen for the qepA gene Additional file 1Table S1. Amplicons were sequence-verified.
The qualitative organic solvent tolerance test used by Wang et al [ 18 ] was employed. Briefly 10 3 4 mid-log phase bacteria were spotted onto glass plates of LB agar and overlaid with hexane, cyclohexane or a [vol:vol] mixtures of these solvents.
Pseudomonas aeruginosa strain PA14 and E. coli MG were used as positive and negative controls respectively. Total RNA was extracted from late log-phase cultures using the Qiagen RNeasy Protect bacterial minikit, chloroquine use and fluoroquinolone resistance dissertation. Following reverse transcription using the Taqman Reverse Transcription Reagents and random hexamers Applied Biosystemsquantitative real-time PCR with SYBR green was performed using primer pairs specific for genes of the components of the efflux pumps or efflux pump regulators using primers of Nishino et al.
The reactions were run on the StepOne Real-time PCR system and performed according to manufacturers recommendations Applied Biosystems. Gene expression levels were compared to the control strain MG, and analyzed using the 2 -ΔΔCt method [ 30 ]. The rrsA gene was used as the normalizing gene. Each reaction was performed in quadruplicate.
fliC PCR-restriction fragment length polymorphism RFLP typing was performed as described by Fields et al [ 31 ]. The fliC gene of test E.
coli strains was amplified using the primers F-FLIC1 and R-FLIC2 Additional file 1Table S1. Amplicons were digested with Rsa I and restriction profiles were discriminated after electrophoresis on 2. PCR primers listed in Additional file 1Table S1 were used to amplify gene fragments from the adk, fumC, gyrB, icd, mdh, purA and recAas described by Wirth chloroquine use and fluoroquinolone resistance dissertation al [ 32 ].
Amplified DNA products were sequenced from both ends. Allele assignments were made at the publicly accessible E. Prescription sheets were obtained with permission from the Obafemi Awolowo University Ile- Ife, Nigeria forand chloroquine use and fluoroquinolone resistance dissertation forand and forand Data from the prescription sheets was collated into a spreadsheet anonymously and then the prescription sheets were returned to the University Health Centre.
Prescriptions for all oral and topical antimicrobials, filled at the institution's Pharmacy were documented and units of measurement were standardized to daily-defined doses were computed using the AB Consumption Calculator tool [ 33 ].
Our previous studies which tracked resistance in fecal E. coli isolates between and found uncommon Due to repeated power cuts in Nigeria, that strain archive is lost and none of those isolates are available for analysis. Employing an identical protocol in we recovered nalidixic acid non-susceptible E. coli from 18 The remaining isolates exhibited low-level nalidixic acid resistance similar to that seen with earlier isolates reported previously [ 21 ].
Quinolone-specific resistance mechanisms in and nalidixic acid-resistant isolates. Inwe recovered nalidixic acid-resistant E. coli from 35
Antimalarial drugs animation: Chloroquine
, time: 1:30Chloroquine: mechanism of drug action and resistance in Plasmodium falciparum
Nov 07, · 1. Background. Chloroquine has been described as one of the most important drugs ever used to treat an infection. This cheap and effective drug was the primary antimalarial worldwide until resistance emerged in the s [].Multiple alleles of a chloroquine resistance transporter gene, pfcrt, and a multidrug resistance transporter gene, pfmdr1, are associated with chloroquine resistance in Cited by: 29 Mechanisms of Fluoroquinolone Resistance In all species studied, mechanisms of fluoroquinolone resistance include one or two of the three main mechanistic categories, alterations in the drug target, and alterations in the permeation of the drug to reach its target. No specific quinolone-modifying or -degrading enzymes have been found Abstract. Quinolone and fluoroquinolone antibiotics are potent, broad-spectrum agents commonly used to treat a range of infections. Resistance to these agents is multifactorial and can be via one or a combination of target-site gene mutations, increased production of multidrug-resistance (MDR) efflux pumps, modifying enzymes, and/or target-protection blogger.com by:
No comments:
Post a Comment